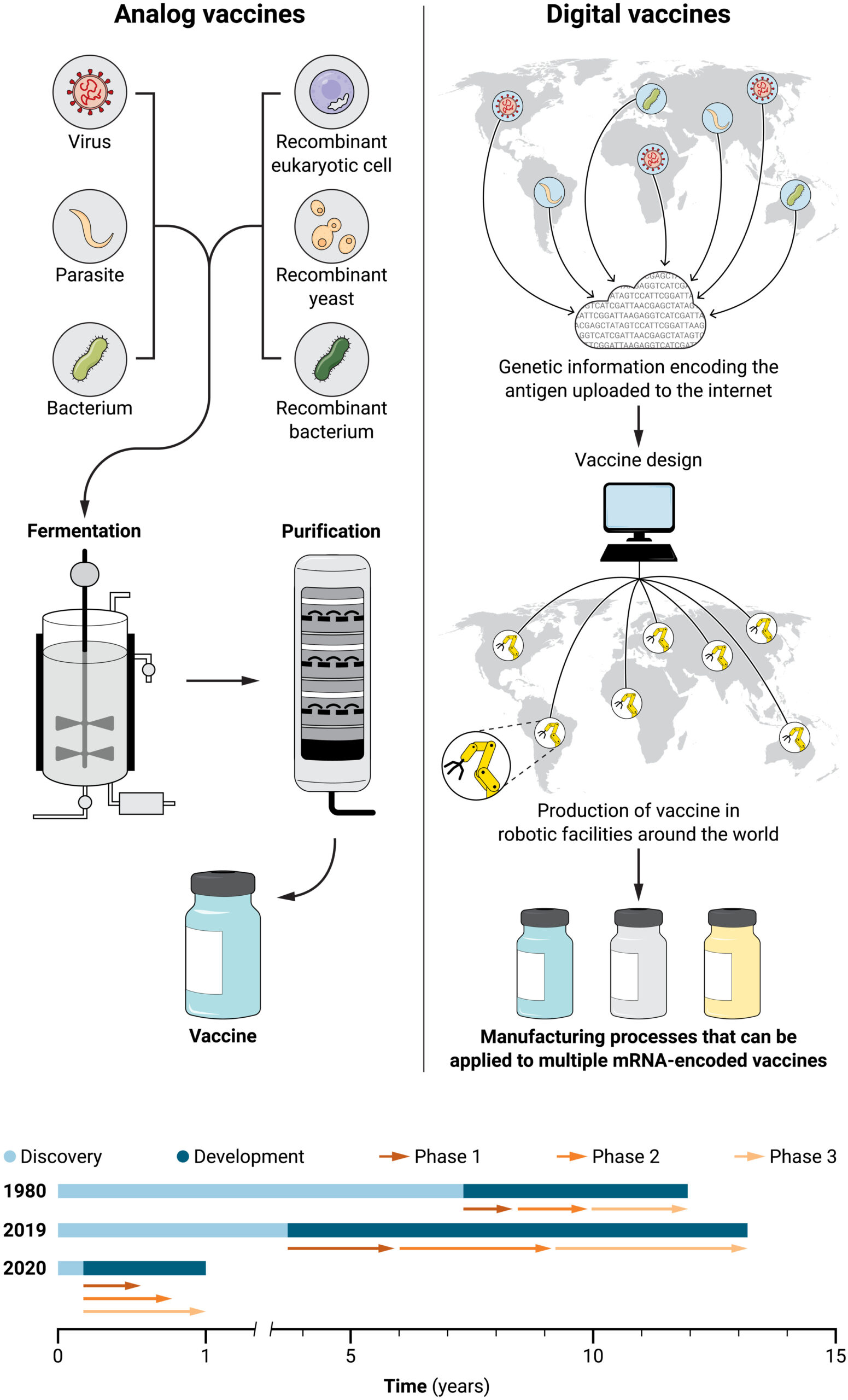
The essence of vaccination consists of exposing the human body to a surrogate of a pathogenic microorganism so that the human immune system can learn how to recognize and fight the foreign invader (
1). Vaccination dates back to ancient China and the Ottoman empire. Smallpox virus, harvested from the pustules of sick people, was inoculated into healthy people with a procedure known as variolation. People were fully protected by this procedure, but the treatment often caused severe disease resulting in death of some of the inoculated individuals. Vaccination in the modern era began when Edward Jenner (
2) made the astute observation that inoculating people with cowpox, a nonvirulent form of the smallpox virus isolated from cows, could educate the immune system to prevent smallpox disease. The new procedure was named “vaccination,” as the word cow is “vacca” in Latin. A century later, Louis Pasteur rationally attenuated pathogens, such as the rabies virus and the bacterium causing chicken cholera, to present nonvirulent surrogates of real pathogens to the host immune system. In the late 19th century, Emil von Behring and Shibasaburo Kitasato (
3) realized that it was sufficient to deliver killed bacteria or even crude bacterial cultures containing diphtheria toxin to elicit serum antibodies that inhibited bacterial replication or neutralized the toxin. New technologies allowed the production of inactivated diphtheria and tetanus toxins, rational attenuation of the
Mycobacterium tuberculosis bacterium to produce the Bacille Calmette-Guérin vaccine, and rational attenuation of the yellow fever virus to yield the 17D vaccine. Killed bacteria have been used in vaccines against typhoid fever and pertussis. The production of vaccines based on surrogate pathogens was accelerated by the discovery that viruses could be grown in chicken eggs or cell cultures. This allowed the development of killed vaccines for influenza; killed and attenuated vaccines against poliomyelitis; and live attenuated vaccines against measles, mumps, rubella, and varicella zoster. Purified components of bacteria such as capsular polysaccharides were then used as vaccines against meningococcus and pneumococcus. With the genetic engineering revolution of the 1980s, recombinant antigens, that is, immunogenic proteins expressed in a different cell rather than the original pathogen, began to be used in vaccines. Indeed, yeast was engineered to express the hepatitis B virus antigen (
4). Other recombinant antigens were produced in yeast;
Escherichia coli; or mammalian, insect, or plant cells, enabling production of vaccines against papillomavirus, meningococcus B,
Borrelia burgdorferi (the bacterial pathogen causing Lyme disease), and herpes zoster. Vaccines using recombinant antigens of respiratory syncytial virus, cytomegalovirus, and coronaviruses have been brought to clinical development. Although recombinant antigens made the design and development of many vaccines possible, they have downsides. For example, discovery is a long and complex process, and scale-up and manufacturing take even longer because new production processes must be set up for each antigen. In many instances, vaccines took 10 to 15 years to be developed and approved (
Fig. 1).